Introduction
Suppose we are given a finite set, , of points in . The set of points is topologically uninteresting. How can we fix this? Yes, you guessed it: we can construct a Vietoris Rips (Rips) complex from the points in ! The Rips Filtration is a specific nested sequence of Rips complexes over . Both provide geometric and topological information about our point set. In this tutorial, we explore this idea and how a Rips complex and filtration can be used to better understand data sets.
Objectives
By the end of this tutorial you should be able to:
- Define Abstract Simplicial Complexes.
- Compute a Rips complex and Rips filtration from a finite set of points in by hand.
- Compute a Rips Filtration from a finite set of points in using the R-TDA package.
- Name a few applications of the Rips complexes and filtrations.
Theory
First, we breifly describe the mathematical foundations behind a Rips Filtration including abstract simplicial complexes, filtrations, and the Rips complex. Computation Topology: An Introduction provides more details on the theory (Edelsbrunner & Harer, 2010).
Triangle Appreciation
We construct a Rips complex from simplices of varying dimensions that are generalizations of triangles of varying dimensions. More specifically, an n-simplex is the smallest convex set of points, where are linearly independent.
An abstract simplicial complex is a finite collection of sets, , that is closed under the subset relation, i.e., if and , then . The elements are the simplices where dimcard. Here, card is the number of points in the set, .
Rips Complex
Let be finite set of points in . Let . The Rips complex of and is the abstract simplicial complex of consisting of all subsets of diameter at most :
where the diameter of a set of points is the maximum distance between any two points in the set.
Geometrically, we can constuct the Rips complex by considering balls of radius , centered at each point in . Whenever balls have pairwise intersections, we add a dimensional simplex. For this tutorial, we use the standard Euclidean distance (unless stated otherwise) to compute a Rips complex.
You may be familiar with contact graphs where the vertices represent a geometric object such as a circle, curve, or polygon, and an edge between two vertices exists if the corresponding two objects intersect. The Rips complex is a generalization of contact graphs.
Here are some examples of Rips complexes:
Note: Many people also define . However, the algorithms used in the R-TDA package use the first definition.
Filtrations
A filtration of a simplicial complex, , is a nested sequence of subcomplexes starting at the empty set and ending with the full simplicial complex, i.e.,
Going back to the Rips complex, we consider to be a free parameter. If we vary , we get different Rips complexes. In many data analysis situations, the value of that best describes the data is unknown or does not exist, so why not look at all of them!? Observe if we increase continuously, then we get a family of nested Rips complexes; the Rips filtration.
Let’s work through an example. Let . We want to compute a Rips filtration on for all .
Observe:
- When , none of the balls of radius intersect and so ) is four points.
- When , the balls of radius centered at and intersect which means we add a 1-simplex between and . Similarly, we add a 1-simplex between and .
- When , no additional balls of radius intersect which means ).
- When , we add a two more 1-simplices between , and .
- When ,
- When , we add two 2-simplices between and .
- When ,
- When , we add a 3-simplex.
- When ,
Applications of Rips Complexes and Filtrations
Now that we have discussed the theory, let’s talk about some of the applications! A Rips filtration is used on point cloud data or more generally on a data set where there is some notion of distance between all data points. A common form of these types of data are location data. For example, coordinates of galaxies in the Universe and GPS coordinates of airports on Earth.
The Rips complex provides information on how close the data points are to each other. Furthermore, the holes in the complex can provide information on where data are not present. Lastly, the topological information given by taking a Rips filtration can be studied using persistent homology (tutorial to come soon!).
Another application is studying coverage in sensor networks. The problem is to look at a set of sensors and find how much coverage the sensors provide. Robert Ghrist and Vin de Silva studied this problem using Rips complexes since the complex can be determined easily from pairwise communication data (Silva & Ghrist, 2007).
Furthermore, Rips complexes can be used in shape reconstruction. Dominque Attali, Andre Lieutier, and David Salinas proved that the Rips complex of a point cloud, , at some threshold has the same homotopy type as the union of balls centered around each point in with radius (Attali, Lieutier, & Salinas, 2013). Rips complexes are nice to use for shape reconstruction since they do not favor a specific type of alignment of the input.
These are just a few of the applications. There are many more!
Computing the Rips Filtration using the R-TDA Package
Toy Example
First, let’s go back to the example where . We compute the Rips filtration using the R-TDA package.
library(TDA) # upload TDA package
S <- cbind(c(0,1,2,3),c(0,3,-1,2)) # write S into R
# Compute Rips Filtration. maxdimension is max dim of homological features
# to be computed, maxscale is the filtration limit.
RipsFilt <- ripsFiltration(S, maxdimension = 2, maxscale = 5,
dist = "euclidean")Great! So how do we interpret the RipsFilt object?
The ripsFiltration function returns a list:
- complx is a list where the element is a vector of the vertices for the simplex.
- values is a vector where the entry is the filtration value for the simplex.
- increasing is a logical variable that states TRUE if the filtration values in the values vector are in increasing order and FALSE if the filtration values are in decreasing order.
- coordinates is a matrix representing the coordinates of the vertices. The row is the coordinate for the vertex. Note the coordinates matrix appears only if the Euclidean distance is used.
See the documentation for this function with this link. Let’s check this out.
RipsFilt$cmplx[[10]] #Access vertices for the 10th simplex in the list## [1] 4 2 1Our output tells us that we have a 2-simplex on vertices . Let’s see at which filtration value this appears.
RipsFilt$values[10] #Access filtration value for the 10th simplex in the list## [1] 3.605551So we have a 2-simplex when . We also found this by hand! Accessing the other complex and filtration value elements verifies the rest of the filtration computation we did earlier.
If we want to see an animation of the Rips filtration, we can run the following code with a plot function. This function has input arguments:
- filt is the Rips filtration object. (So, Ripsfilt from above)
- animate is a logical variable stating TRUE if you want an animation of the filtration or FALSE if you do not want an animation.
- thresh is the threshold value for the Rips complex that will be plotted if animate = FALSE.
- fn_out is the name of the file for the animation if animate = TRUE.
- l_out is the number of levels of radii used in the animation between 0 and 1.1 times the max distance between any two points if animate = TRUE.
Note this function only works if the data set is two dimensional and the Euclidean distance is used.
library(plotrix) # packages for plot function
library(animation)
class(RipsFilt) <- c(class(RipsFilt), "rips_filt")
plot(RipsFilt, animate = TRUE, fn_out = 'rips_example.gif',
lout = 80, interval = 0.2)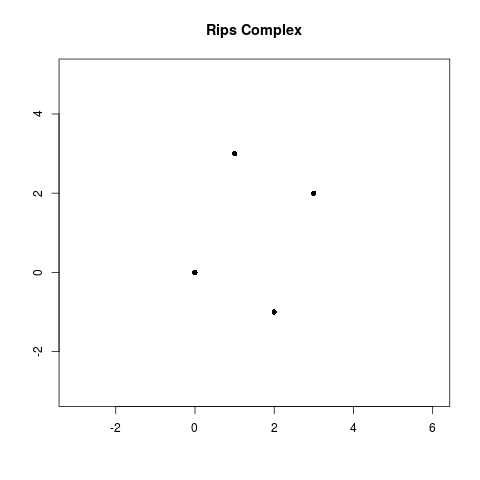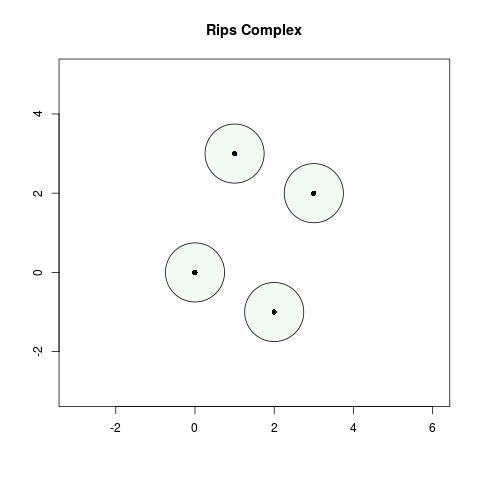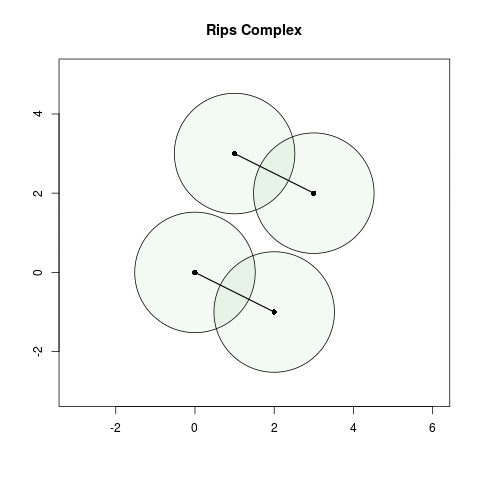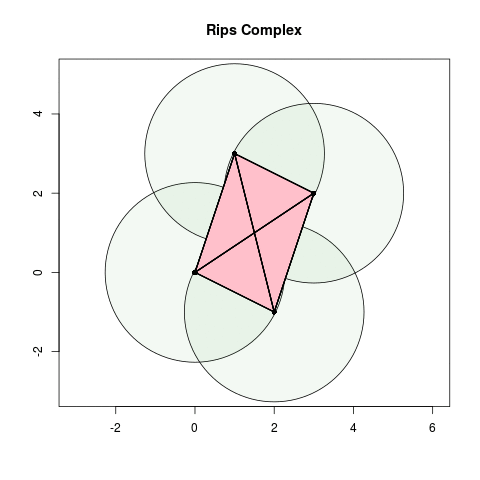
Furthermore, we can look at the Rips complex at a threshold of , by adding the argument, thresh = and changing animate = FALSE to animate = TRUE.
plot(RipsFilt, thresh = sqrt(14), animate = FALSE)Non-Euclidean Distance Example
Now we compute a Rips filtration on , but this time using the Manhattan distance or norm. Recall, that for points, and , the Manhattan distance, is defined as:
In order to compute the Rips filtration on , we need to create a distance matrix in R. We can do this using the following code:
DistMatrix <- dist(S, method = "manhattan", diag = TRUE, upper = TRUE, p = 2)When we use the ripsFiltration function, we write DistMatrix as the first argument instead of . Additionally, the fourth argument will change to dist = “arbitrary” to indicate that we are giving our own distance metric.
RipsFiltManhattan <- ripsFiltration(DistMatrix, maxdimension = 2,
maxscale = 5, dist = "arbitrary")This time when we compute the Rips complex with , we get the following complex:
library(squash) # package for drawing diamonds
lim <- rep(c(-3, 5), 2)
plot(NULL, type = "n", xlim = lim[1:2], ylim = lim[3:4],
main = "Rips Complex with Manhattan Distance for r = sqrt(14)",
xlab = "", ylab = "")
for(j in 1:length(S[, 1])) {
diamond(x = S[j, 1], y = S[j, 2], radius = sqrt(14) / 2, col = rgb(0.1, 0.6, 0.1, 0.05))
}
for (idx in seq(along = RipsFiltManhattan[["cmplx"]][RipsFiltManhattan$values < sqrt(14)])) {
polygon(S[RipsFiltManhattan[["cmplx"]][[idx]], , drop = FALSE],
col = "pink", border = NA, xlim = lim[1:2], ylim = lim[3:4])
}
for (idx in seq(along = RipsFiltManhattan[["cmplx"]][RipsFiltManhattan$values < sqrt(14)])) {
polygon(S[RipsFiltManhattan[["cmplx"]][[idx]], , drop = FALSE],
col = NULL, xlim = lim[1:2], ylim = lim[3:4])
}
points(S, pch = 16)Hence with the Manhattan distance is a simplicial complex with two edges, while with the Euclidean distance is a simplicial complex with two 2-simplices.
Sightings of Bigfoot in Montana
Lastly, we work through an example with real data! The Bigfoot Field Researchers Organization (BFRO) is the only scientific research organization studying the Bigfoot/Sasquatch mystery. They publically provide all Bigfoot sighting reports on their website. We will compute the Rips filtration for Bigfoot sightings in Montana with the hope of finding the where Bigfoot is and is not roaming. Finding the deadzones where Bigfoot is not present could help keep hikers safe from potential encounters.
#Latitude coordinates of Bigfoot sightings in MT
latitude <- c(45.31278,46.05292,45.97540,47.62167,48.03278,47.70667,
46.02962,47.66724,46.01833,46.97694,46.76389,45.12266,
48.27011,46.76472,46.18694,48.36139,45.22417,47.60413,
47.17833,45.28074,46.48392,47.57549,45.79220,45.61331,
48.68000,46.30766,47.07444,47.04333,47.07954,46.83285,
47.12808,46.59692,47.03222,47.06032,46.75889,46.96508,
46.52520,45.66722,45.82285,45.02620,45.37050)
#Longitude coordinates of Bigfoot sightings in MT
longitude <- c(-109.64490,-112.57100,-112.45540,-115.39030,-105.86420,
-104.19220,-113.11980,-108.68730,-110.33080,-113.83810,
-110.77810,-111.56300,-115.64180,-111.88610,-113.69250,
-113.12360,-111.12500,-113.75930,-111.97440,-109.55280,
-111.26440,-114.2650,-111.36370,-110.78970,-113.82000,
-113.14240,-111.96250,-112.70310,-112.56490,-112.70200,
-111.90940,-110.62680,-114.09060,-114.07170,-114.08000,
-114.07250,-113.59110,-110.80690,-114.01250,-106.83310,
-112.97070)
#GPS coordinates of Bigfoot sightings in MT
BigfootSightingsMT <- cbind(longitude, latitude)We can plot the data using the following code.
mt_range <- map("state", "Montana", plot = FALSE)$range
xrange <- diff(mt_range[1:2])
yrange <- diff(mt_range[3:4])
plot(BigfootSightingsMT[,1],BigfootSightingsMT[,2], xlab = "Latitude",
ylab = "Longitude", main = "GPS coordinates of Bigfoot sightings
in Montana", xlim = c(mt_range[1] - .1*xrange, mt_range[2] + .1*xrange),
ylim = c(mt_range[3] - .1 * yrange, mt_range[4] + .1 * yrange), pch=16)
map("state", "Montana", add = TRUE)Now, let’s compute the filtration and visualize it with an animation. In order to speed up the computation, we will set maxscale = 2.5 and maxdimension = 3.
BigfootRipsFilt <- ripsFiltration(BigfootSightingsMT, maxdimension = 3,
maxscale = 1.5, dist = "euclidean")
class(BigfootRipsFilt) <- c(class(BigfootRipsFilt), "rips_filt")
plot_rips_bigfoot(BigfootRipsFilt, animate = TRUE, fn_out = 'bigfoot.gif',
lout = 120, interval = 0.3, ani.height = 240, ani.width = 240*2)Furthermore, we can look at the Rips complex at a threshold of 0.8.
plot_rips_bigfoot(BigfootRipsFilt, thresh = 0.8, animate = FALSE)As we watch the animation, we see that the Rips filtration captures geometric properties about the data set since simplices quickly arise in the filtraion in the locations that reported the most bigfoot sightings. Furthermore, we see the filtration also reveals some topological information about the data set. In particular, at a filtration value of 0.698812, five edges that form a cycle appear around the GPS coordinate (-111, 46). Furthermore, this cycle persists until the filtration value of 0.94698369. Considering the amount of land that is covered in 1 longitude/latitude unit, this cycle could tell us something meaningful about our data. Perhaps this hole is a deadzone where Bigfoot has yet to be found, or maybe this area is inaccessible to humans and that is why there has not been sightings. Regardless of the answer, this cycle provides us guidance for the next step to take to solve the Bigfoot mystery!
References
- Edelsbrunner, H., & Harer, J. (2010). Computational Topology - an Introduction. (pp. 1–241). American Mathematical Society.
- Silva, V. de, & Ghrist, R. (2007). Coverage in Sensor Networks via Persistent Homology. Algebraic & Geometric Topology, 7. https://doi.org/10.2140/agt.2007.7.339
- Attali, D., Lieutier, A., & Salinas, D. (2013). Vietoris–Rips Complexes also Provide Topologically Correct Reconstructions of Sampled Shapes. Computational Geometry, 46(4), 448–465. https://doi.org/https://doi.org/10.1016/j.comgeo.2012.02.009